Advanced topics in Deep Learning, ESPCI
This 3rd-year course is a follow up of an introduction to deep-learning in 2nd year. It covers some advanced topics in deep-learning with guest lectures and lab sessions in pytorch (python module). The plenary session include exercises ( or TD).
News
- First course: January the 9th. Early in the morning !
Expected schedules
It starts in january 2023 (the 9th). The course is scheduled on monday, starting at 8:30 in the morning.
- 9/01, course: introduction, reminder and exercises
- 16/01, course:
- Reminder - 2
- Adversarial attack and defense
- Pytorch
- 23/01, lab session 1
- Refresher on pytorch
- Convolution on CIFAR10
- 30/01, lab session 2
- ResNet
- Adversarial attacks and defense
- 06/02, course:
- Sequence and convolution 1D
- Recurrent Network
- Optimization
- Project
- 13/02, lab session 3
- Recurrent Network
- 20/02, course:
- Generative models (TBC)
- 13/03, guest course: Applications to Biology
Vaitea Opuu
- 20/03, guest course: Deep Learning for Astro-Physics
David Cornu
- 27/03, guest course: Machine Learning, Physics and Fluid Mechanics
Lionel Mathelin
python and Notebooks: how to
We will use python 3, pytorch and notebooks. If you need to work with own computer, there are 2 ways:
- install anaconda 3 on your computer: see this page.
- use colab with a google account (the easiest, nothing todo)
To use files stored on your google drive you can add in your colab notebook:
from google.colab import drive drive.mount('/content/gdrive') # in my drive, I have a directory "Colab Notebooks" # the dataset is uploaded there root_path = 'gdrive/My Drive/Colab Notebooks/'
If you are not familiar with python notebooks, see this page.
Projects
Here, you can find a list of possible projects. Feel free to interact with me. For some of them, just ask me the data, otherwise a link is provided. Of course, you can also propose a project. This section is under construction and maybe some projects will be added as soon as I will have more feedbacks from my colleagues.
- Integrating Deep Learning with Microfluidics for Biophysical Classification of Sickle Red Blood Cells
- Cell Tracking challenge: Segmenting and tracking moving cells in time-lapse video sequences is a challenging task, required for many applications in both scientific and industrial settings.
- COVID Forecasting
- Predicting clustered weather patterns: A test case for applications of convolutional neural networks to spatio-temporal climate data.
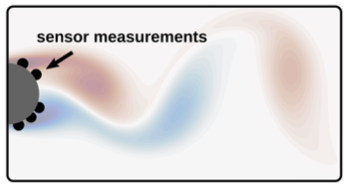
- Reconstruction of the vorticity field of a flow behind a cylinder from a handful sensors on the cylinder surface
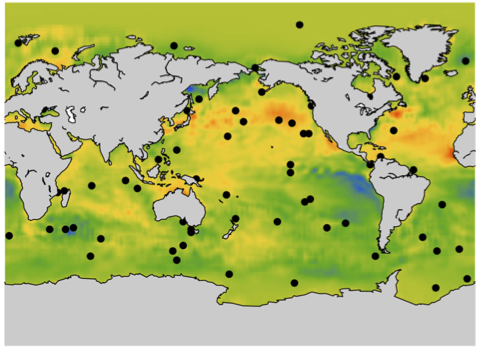
- The mean sea surface temperature reconstruction from weekly sea surface temperatures for the last 26 years. You can also have access to other measures. For this project, you can read the paper associated with the previous project or also look at this recent paper.
- Chaos as an interpretable benchmark for forecasting and data-driven modelling
- Predicting the sequence specificities of DNA- and RNA-binding proteins. We can use datasets from Deep-bind.
- Deep sequence models for protein classification: there is a recent paper on this topic and data can be available. We can try different models (maybe simpler) for the same task.
- Chemistry: Predict the standard density of pure fluids, using a newly compiled database. From SMILES description, how can we predict density ? Ask me for the data and tools.
- Classify sleep and arousal stages from physiological signals including: electroencephalography (EEG), electrooculography (EOG), electromyography (EMG), electrocardiology (EKG), and oxygen saturation (SaO2). See the challenge page for more details
- Classify, from a single short ECG lead recording (between 30 s and 60 s in length), whether the recording shows normal sinus rhythm, atrial fibrillation (AF), an alternative rhythm, or is too noisy to be classified: The challenge page. Other datasets can be nice:
- Using seismic signals to predict the timing of laboratory earthquakes.
- Quantum-mechanical molecular energies prediction from the raw molecular geometry: see the QM7 database.
- Classify Molecule polarization: the data comes from time-lapse fluorescence microscopy images of the bacterium Pseudomonas fluorescens SBW25. Each image is an individual bacterial cell. These bacteria produce a molecule called pyoverdin which is naturally fluorescent, so the images show the distribution of this molecule inside the cells. We have discovered that there are two distribution patterns of this molecule: homogeneous, or accumulated at the cell pole ("polarized").
- Jet Flavor Classification in High-Energy Physics: http://mlphysics.ics.uci.edu/
- Recognize decays in real high energy physics experiment: https://www.kaggle.com/c/beta-beta-decay-identification/data
- A project associated with the course of statistical physics on the XY-model
- Simple Recurrent network for language modelling, coupled with adaptive stepsize
- 1-Lipschitz recurrent networks : language model and DNA sequence analysis